Hi
When running split_multi_hts on a matrixtable I am finding GQs are being changed, can someone explain this behaviour to me.
I am happy to provide VCF and jupyter notebook for the below if you can advise me on how to share them with you (vcf is 372M).
Details follow:
Running Hail version 0.2.134-952ae203dbbe
I have a WGS VCF for 5 samples which has been variant called using DeepVariant and GLnexus.
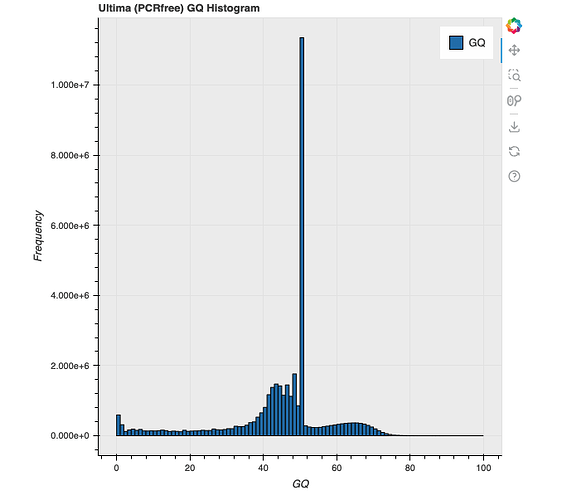
I first noticed a problem when producing histograms of GQ using the following command
p = hl.plot.histogram(pcrfree_mt.GQ, range=(0,100), title='Ultima (PCRfree) GQ Histogram', legend='GQ')
The histrogram showed roughly the expected distribution
I then split the multiallelics
pcrfree_mt_split = hl.split_multi_hts(pcrfree_mt)
plotting the histogram after splitting showed > 9 million variants with GQ ~98/99, which were not there in the dataset prior to splitting
The following is a screenshot from the output of mt.summarize.entries() from my unsplit data - this shows a maximum GQ of 90
This is a screenshot of mt.summarize.entries() from the data after split_multi_hts, showing the maximum GQ is now 99
There are now 9723159 entries with GQ of > 90. I checked a handful against the raw VCF and the GQ has definitely been altered.
The entries which now have a GQ of >90 include entries which were split by split_multi_hts and entries which were not split.
pcrfree_mt_split_highGQ = pcrfree_mt_split_highGQ.filter_rows(pcrfree_mt_split_highGQ.variant_qc.n_called > 1)
pcrfree_mt_split_highGQ.aggregate_rows(hl.agg.counter(pcrfree_mt_split_highGQ.was_split))
{False: 2849457, True: 294118}
I hope someone can help
Best wishes
Ruth